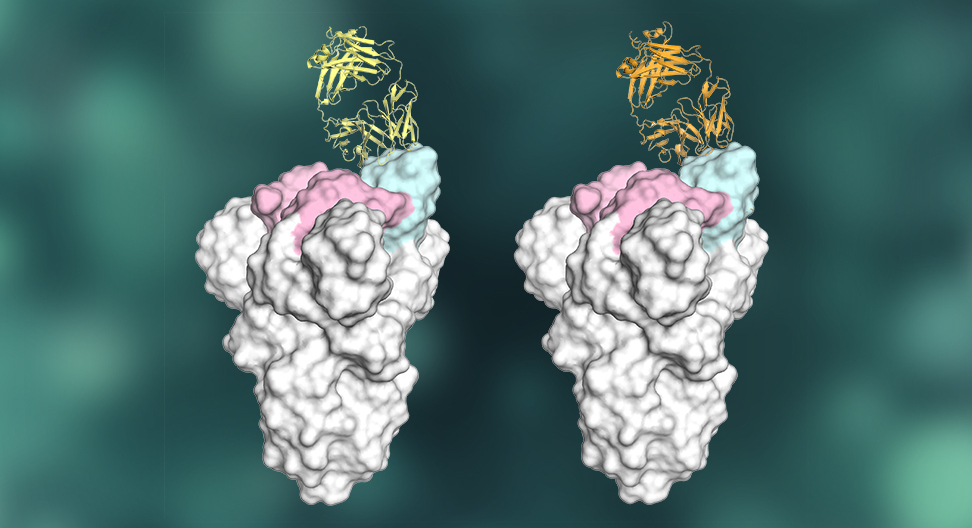
Many anti-SARS-CoV-2 antibodies are produced by a common antibody gene. X-ray crystallography revealed how specific features of these antibodies (two of which are depicted here in yellow and orange, attached to the virus) enable potent recognition of the virus's spike protein. Graphic by Meng Yuan, Hejun Liu and Nicholas Wu in the Wilson lab.
A team led by scientists at Scripps Research has discovered a common molecular feature found in many of the human antibodies that neutralize SARS-CoV-2, the coronavirus that causes COVID-19.
The scientists, whose study appears July 13 in Science, reviewed data on nearly 300 anti-SARS-CoV-2 antibodies that their labs and others have found in convalescent COVID-19 patients over the past few months. They noted that a subset of these antibodies is particularly powerful at neutralizing the virus—and these potent antibodies are all encoded, in part, by the same antibody gene, IGHV3-53.
The scientists used a powerful tool known as X-ray crystallography to image two of these antibodies attached to their target site on SARS-CoV-2. The resulting atomic-structure details of this interaction should be useful to vaccine designers, as well as to scientists hoping to develop antiviral drugs targeting the same site on SARS-CoV-2.
Prior research suggests that antibodies encoded by IGHV3-53 are generally present, at least in small numbers, in healthy people’s blood. The results therefore offer hope that using a vaccine to boost levels of these ever-present antibodies will protect adequately against the virus.
“This type of antibody has been isolated frequently in studies of COVID-19 patients, and we can now understand the structural basis for its interaction with SARS-CoV-2,” says the study’s senior author Ian Wilson, DPhil, Hansen Professor of Structural Biology and Chair of the Department of Integrative Structural and Computational Biology at Scripps Research.
“This study provides important inspiration for effective COVID-19 vaccine design,” says co-author Dennis Burton, PhD, professor and co-chair of the Department of Immunology and Microbiology at Scripps Research.
The research was a collaboration chiefly involving the Wilson and Burton labs, and the Scripps Research-based Neutralizing Antibody Center of IAVI, a prominent non-profit vaccine research organization.
SARS-CoV-2 so far has infected more than 12 million people around the world and killed more than 500,000, in addition to causing widespread socioeconomic disruption and damage. Developing an effective vaccine to stop the pandemic is currently the world’s top public health priority.
Although several potential vaccines are already in clinical trials, scientists don’t yet have a full understanding of the molecular features that would define a protective antibody response. In the new study, the scientists took a big step toward that goal.
The team started by analyzing 294 different SARS-CoV-2-neutralizing antibodies that have been isolated from COVID-19 patients’ blood in studies over the past few months. Antibodies are Y-shaped proteins made in immune cells called B-cells. Each B-cell makes a specific antibody type, or clone, which is encoded by a unique combination of antibody genes in the cell. The scientists found that an antibody gene called IGHV3-53 was the most common of the genes for the 294 antibodies, encoding about 10 percent of them.
The scientists also noted that the IGHV3-53-encoded antibodies in their study contain an unusually short variant of the CDR H3 loop, normally a key target-binding element. These antibodies are nevertheless very potent against SARS-CoV-2 when compared to other antibodies not encoded by IGHV3-53.
A powerful response right off the bat
The IGHV3-53 antibodies had yet another property suggesting that boosting their numbers would be a good and achievable aim for a SARS-CoV-2 vaccine: They appeared to have mutated only minimally from the original versions that would be circulating, initially in small numbers, in the blood of healthy people.
Normally, when activated by an encounter with a virus to which they fit, B-cells will start proliferating and also mutating parts of their antibody genes, in order to generate new B-cells whose antibodies fit the viral target even better. The more mutations needed for this “affinity maturation” process to generate virus-neutralizing antibodies, the harder it can be to induce this same process with a vaccine.
Fortunately, the IGHV3-53 antibodies found in the study seemed to have undergone little or no affinity maturation and yet were already very potent at neutralizing the virus—which hints that a vaccine may be able to induce a protective response from these potent neutralizers relatively easily.
“Coronaviruses have been around for hundreds to thousands of years, and one can imagine that our immune system has evolved in such a way that we carry antibodies like these that can make a powerful response right off the bat, so to speak” Wilson says.
Map for vaccine-makers, gauge for clinical trials
Wilson’s team used high-resolution X-ray crystallography to image two different IGHV3-53 antibodies bound to their target on SARS-CoV-2. This target, known as the receptor binding site, is a crucial structure on the viral “spike” protein that normally connects to a receptor on human cells to begin the process of cell infection. Many of the antibodies that neutralize SARS-CoV-2 appear to do so by blocking this virus-receptor connection.
“We were able to reveal unique structural features of these IGHV3-53-encoded antibodies—features that facilitate their high binding affinity and their specificity for the SARS-CoV-2 receptor binding site,” says co-first author Meng Yuan, PhD, a postdoctoral research associate in the Wilson lab.
The detailed atomic-scale structural data should be of interest to vaccine designers and drug developers. Moreover, the researchers say, the identification of IGHV3-53-encoded antibodies as key elements of the immune response to COVID-19 suggests that levels of these antibodies might be useful as an indirect marker of success in ongoing and future vaccine trials.
“Structural basis of a shared antibody response to SARS-CoV-2” was written by first authors Meng Yuan, Hejun Liu, and Nicholas Wu, all of the Wilson lab; and by Chang-Chun Lee, Xueyong Zhu, Fangzhu Zhao, Deli Huang, Wenli Yu, Yuanzi Hua, Henry Tien, Thomas Rogers, Dennis Burton, and Ian Wilson, all of Scripps Research; and Elise Landais, Devin Sok and Joseph Jardine of IAVI.
The research was supported by the National Institutes of Health (K99 AI139445, UM1 AI44462), the Bill and Melinda Gates Foundation (OPP1170236) and IAVI.


