 Fig. Many serious diseases are caused by changes in the protein structure. Proteogenomic technology can be used to identify mutant proteins.
Fig. Many serious diseases are caused by changes in the protein structure. Proteogenomic technology can be used to identify mutant proteins.
Proteomics and Big Data
It seems as if the term «big data» is relatively new. However, it is impossible to imagine any science that would not use large amounts of data. In biology, big data enables scientists to, on the one hand, conduct large-scale experiments and extract more useful data from biological material. On the other hand, it is becoming increasingly difficult to reveal important patterns of high specificity within the large amount of information. To cope with this problem, scientists are increasingly focusing on developing complex algorithms and/or the workflows for filtering and analyzing the data.
Proteomics — the large-scale study of proteins of cells and entire organisms — is no exception. Generally, proteins, peptides, and their fragments can be analyzed using mass spectrometry. Mass spectrometry provides peptide fragmentation information specific to the amino acid sequence and, thus, allows scientists to identify proteins present in the original sample. A number of algorithms called a search engine are currently available for protein identification. These algorithms take the peptide fragmentation patterns provided by mass spectrometry, match them with a protein database and return the list of proteins corresponding to the experimental data.
However, this approach is not entirely suitable for proteins that are not encoded in a reference genome. If a mutant protein from a cancer cell does not present in the search database, then the so-called variant peptide corresponding to the mutated part of the protein would not be identified. This is where proteogenomics comes in — a rapidly growing area of biological research at the intersection of genomics and proteomics. Variant peptides identified using the proteogenomic approach provide invaluable information for gene annotation — information which is difficult or impossible to ascertain using standard annotation methods.
Expansion of the protein database
In their paper, the Russian scientists describe a workflow for searching variant peptides from mutant proteins enabling them to compare the mass spectrometry results of different groups and laboratories for unambiguous marking of cancer mutations. The effectiveness of their approach has been tested using HEK-293 cells. HEK-293 (Human Embryonic Kidney 293) cells are a specific cell line originally derived from human embryonic kidney cells grown in a tissue culture. HEK-293 cells have been widely used in cell biology research for many years because of their reliable growth and propensity for transfection.
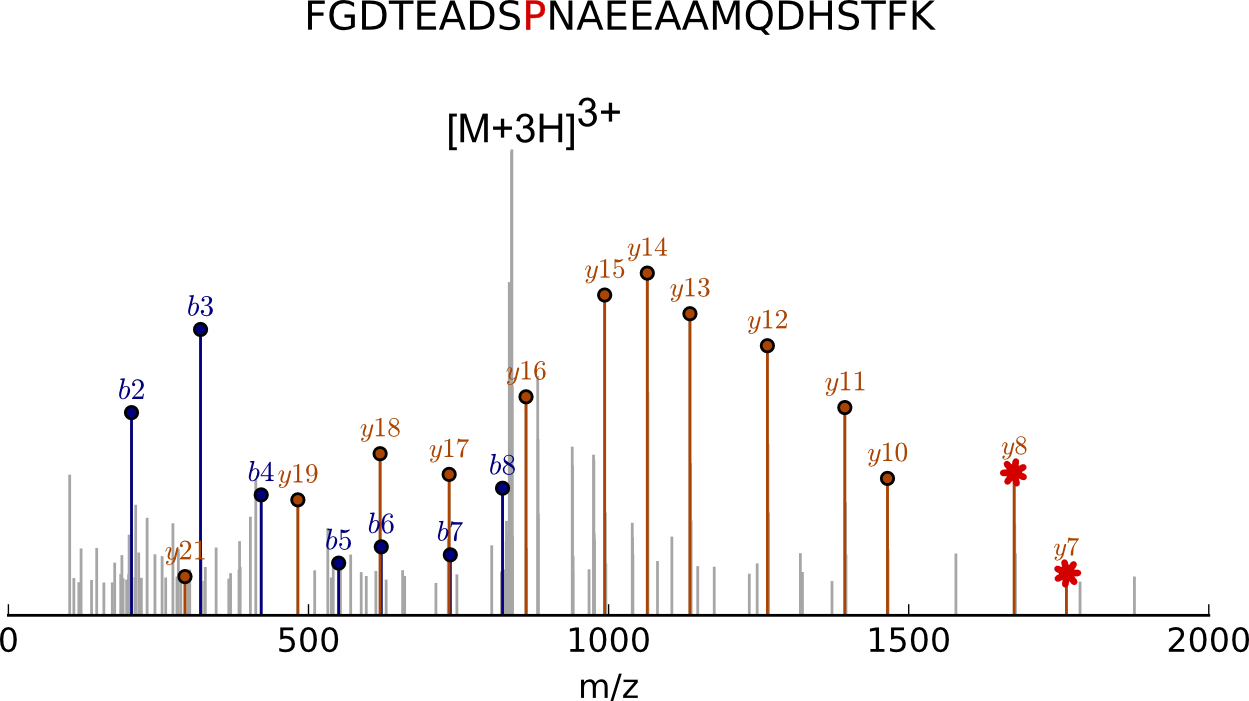
Fig. Mass spectrum of peptide fragments [from a Dmx‑like 2 protein] showing a point mutation. The red indicates the position where an amino acid residue substitution (P for S) occurred. It also marks the peaks in the mass spectrum that confirm this substitution.
In addition to their own experimental data, the researchers used the mass spectrometry results from two recent studies analysing HEK-293 cell proteomes. They generated the so-called customized database for proteogenomic analysis based on exome sequencing of HEK-293 cells. An exome is formed by exons (part of a gene that codes an amino acid sequence). As a result, the customized protein database now has 1336 sequences of mutant proteins in addition to the reference database of human proteins. This simply means that the protein «dictionary» has grown. Without this improvement it would be impossible to find the «wrong» mutant proteins. A cancer cell mutates more often than a regular cell, which is why known differences between proteins in cancer and «reference» cells will help scientists to find out more about tumor cells.
With the mass spectrometry data available from two previous studies and the own experimental results, the Russian scientists identified peptides and the corresponding proteins contained in the cell. Using the proteogenomic analysis with an expanded peptide database, the authors discovered 113 unique variant peptide sequences in HEK-293 cells referring to the exons of 103 genes.
Some of the mutations discovered had previously been proven to be connected with different types of cancer. These mutant proteins could possibly facilitate the survival and multiplication of the cells. In particular, one of the variants identified is related to the p53 protein which is known to suppress the malignant transformations.
«Our approach may be used to search for cancer-associated mutations based on proteomic analysis. This will help in studying the protein expressions in tumors and provide further basis for developing drugs targeting the mutant proteins produced in tumor cells," says Dr. Michael Gorshkov, one of the collaborators in the project, the Head of the Laboratory of Physical and Chemical Methods for Structure Analysis at the Institute for Energy Problems of Chemical Physics, and a member of MIPT’s Department of Chemical Physics.
Source:
https://mipt.ru/english/news/peptide_mutants_help_identify_vulnerability_in_tumor_cells




