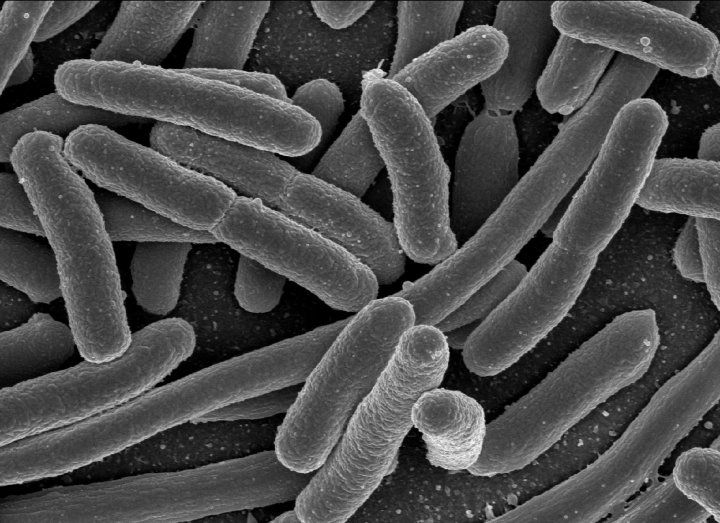
 In the landmark study, published this month in the journal Cell Reports, the researchers were able to remove chunks of genes from the Escherichiacoli bacteria living in the guts of mice and change the overall makeup of the bacterial communities populating their digestive systems.
In the landmark study, published this month in the journal Cell Reports, the researchers were able to remove chunks of genes from the Escherichiacoli bacteria living in the guts of mice and change the overall makeup of the bacterial communities populating their digestive systems.Escherichia coli (E. Coli) bacteria.
“We’ve demonstrated the first stable gene edit within the gut microbiome of a mammal,” said Peter Turnbaugh, PhD, a professor of Microbiology and Immunology. “This is the starting point for trying to engineer bacteria within the gut.”
Currently, researchers and medical practitioners wanting to alter the gut microbe have extremely limited options. Bacterial food poisoning and similar issues can be treated with broad-spectrum antibiotics, for example, but such medication winds up killing off many “good” microbes along with the bad. Fecal transplants, too, have been used in the hopes of reseeding a healthy microbiome in patients with certain infections and gastrointestinal diseases. But physicians can’t be sure that the introduced microbes will manage to outcompete the patient’s existing bacterial communities, meaning the treatment isn’t always successful.
Being able to alter the DNA of microbes already in the gut can allow us to study the microbiome in a more controlled way than we’ve been able to before.
PETER TURNBAUGH, PHD, PROFESSOR OF MICROBIOLOGY AND IMMUNOLOGY
Modifying bacteria already thriving within the digestive system will therefore be a key approach in the research and treatment of microbiome-related health issues in the future, says Turnbaugh. Directly altering the genomes of microbes in the gut would introduce a level of precision to microbiome treatments that hasn’t yet been possible.
“Being able to alter the DNA of microbes already in the gut can allow us to study the microbiome in a more controlled way than we’ve been able to before,” said Turnbaugh. “It really gives us a chance to ask important questions about health and disease.”
Turnbaugh’s study focused on E. coli, a bacterium that occurs naturally in the gut, but which harbors a bad reputation because certain strains can cause food poisoning. One useful application of precision gene editing in the gut microbiome would be to target harmful strains of E. coli while leaving helpful ones undisturbed.
For this study, the researchers wanted to know if they could use the gene editing tool to target and kill one strain of E. coli while leaving another alone. Turnbaugh's team used a virus, called M13, to inject a CRISPR-Cas9 system into the cells of a specific strain of E. coli, where it went to work chopping out segments of DNA.
The results were dramatic. Before the CRISPR-Cas9 system was introduced, the targeted strain was more prominent in fecal samples collected from the mice in the experiment. After gene editing, however, the targeted strain quickly began to disappear. After two weeks, it represented only one percent of the monitored cell population.
A key to the study’s success was the use of an engineered form of M13, a virus that naturally attacks E. coli but doesn’t typically survive well inside the digestive system. To solve that problem, Turnbaugh and his colleagues spliced an antibiotic resistance gene into the DNA that M13 would deliver to the cells it infected, allowing the virus – and the CRISPR-Cas9 system it carried – to spread more easily.
Turnbaugh envisions that a similar approach could someday be used to promote the growth of “good” gut bacteria in humans. For example, if researchers edited genes in certain bacterial strains to allow the bacteria to feed on rare nutrients, a person could gain some control over the mix of microbes flourishing in their gut by simply adding high amounts of those nutrients to their diet. First, he said, researchers will need to expand the list of viruses in their toolkit and experiment with how altering individual members of the microbiome affects the bacteria population overall.
“The dream is that you could just choose which specific strains in your gut – or even just individual genes – you want to promote or take out,” Turnbaugh said. “We’re really excited about how far we were able to push this in E. coli. Hopefully it will lead to similar tools for other members of the gut microbiota.”
By Lindzi Wessel
ucsf.edu


