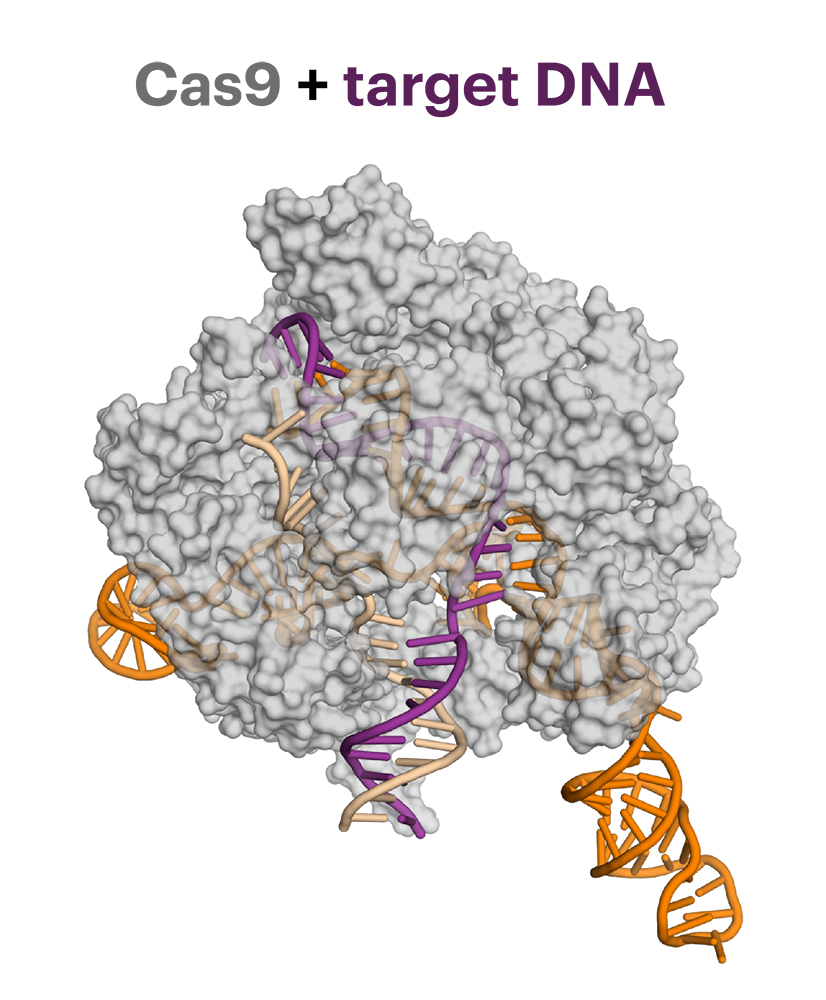
The Cas9 protein gloms onto a targeted piece of DNA (purple) before cutting it. Bacteria developed Cas9 as a weapon to kill viruses by snipping their DNA, but some viruses came up with a defense, called anti-CRISPR, that inactivates Cas9. The guide RNA (orange) directs Cas9 to its target. (Fuguo Jiang image)
In a study reported online this week in the journal Science Advances, researchers from UC Berkeley and UC San Francisco show that recently discovered anti-CRISPR proteins decrease off-target effects by as much as a factor of four, acting like a kill switch to disable CRISPR-Cas9 after it’s done its job.
The study demonstrated that one particular anti-CRISPR protein called AcrIIA4 reduced by four-fold the off-target effects of a CRISPR-Cas9 molecule that uses a guide RNA to find, snip and replace the mutated hemoglobin gene responsible for sickle cell disease. It does this without significantly reducing the desired on-target gene-editing.
“Unexpected mutations can arise as a result of off-target gene editing, but our paper — like many others — shows that off-target effects can be modulated and it is not as serious as people might think,” said UC Berkeley postdoctoral fellow Jiyung Jenny Shin, from the lab of Jacob Corn at the Innovative Genomics Institute and one of three first authors of the paper.
In her experiments on human cells in culture, Shin found that delivering CRISPR-Cas9 and then, several hours later, the anti-CRISPR protein, was the most effective way to reduce off-target effects. The protein mimics DNA, glomming onto Cas9, the enzyme that actually cuts the double-stranded DNA, and preventing further cutting.
“Even after six hours of effective CRISPR, inserting anti-CRISPR decreases off-target effects by more than two-fold compared to on-target effects,” Shin said. “Therapeutically, you could treat a patient with CRISPR first, and then treat with anti-CRISPR at a later time and decrease off-target effects.”
The researcher who discovered AcrIIA4, Joseph Bondy-Denomy of UC San Francisco, foresees these anti-CRISPR proteins becoming a standard part of CRISPR gene therapy, given along with CRISPR-Cas9 to disable gene editing after a fixed period of time to prevent random off-target cutting.
“This Cas9 inhibitor could be encoded on the same piece of DNA as Cas9, for example, precisely timed to turn Cas9 off after the gene editing is done, instead of letting Cas9 linger in the cell and risk off-target effects,” said Bondy-Denomy, who is also a co-author of the paper.
Anti-CRISPR binding
The team included researchers in the lab of Jennifer Doudna, one of the inventors of CRISPR-Cas9 gene editing, who determined how the anti-CRISPR protein binds to the CRISPR-Cas9 complex. Using cryo-electron microscopy, they found that anti-CRISPR essentially mimics DNA, tricking CRISPR-Cas9 into binding with it, and then never letting go.

The anti-CRISPR protein (red on right) mimics DNA, binding in the site where the cutting enzyme Cas9 typically grips the target DNA (left) before it cuts. But the anti-CRISPR protein doesn’t let go, essentially killing Cas9’s gene editing ability. (Fuguo Jiang image?
The CRISPR inhibitor targets a spot on the Cas9 protein that is so essential for Cas9’s function that it cannot operate to cut DNA when it’s bound by the anti-CRISPR.
Last year, Bondy-Denomy reported finding four anti-CRISPR proteins used by attacking viruses to inactivate the version of the Cas9 protein found in the bacterium Listeria monocytogenes. Two of these also inhibited the Cas9 protein most commonly used by researchers, which is adapted from the bacterium Streptococcus pyogenes and is referred to as SpyCas9. Another team found three other anti-CRISPR proteins that work against a different but promising Cas9 protein adapted from the bacterium Neisseria meningitidis.
The current study looked at the effect of one of the proteins from Listeria, AcrIIA4, on SpyCas9 loaded with a guide RNA that homes in on complementary DNA to bind and cut.
Research at UC Berkeley and elsewhere suggests that CRISPR-Cas9 constantly feints with the cell’s DNA repair system: as the enzyme cuts at its target site, the cell repairs the DNA, and CRISPR-Cas9 cuts again, repeating this vicious cycle until a mutation arises in the DNA that prevents enzyme binding, at which point the CRISPR-Cas9 molecule moves on to find another binding site.
The current work from the Corn and Doudna labs now suggests that adding an anti-CRISPR after Cas9 has successfully edited a target gene would prevent unintended damage to other portions of a genome.
“The ability to turn Cas9 gene editing off is just as important as the ability to turn it on,” said Corn, scientific director for biomedicine of the IGI and a UC Berkeley assistant adjunct professor of molecular and cell biology. “Imagine if you had an electric razor with no off-switch! For eventual therapeutic applications, it is critical to be able to precisely control when and where gene editing is active. The anti-CRISPR proteins offer opportunities to completely turn off Cas9 as well as fine-tune its activity.”
“Jenny’s data suggests that there is an ideal time window for letting Cas9 do its job and then turning it off after that amount of time has passed,” Bondy-Denomy said. “We can actually use the anti-CRISPR proteins as tools to figure out what that time window is, that is, for any one cell type with any one guide RNA sequence, how long we want Cas9 to be active in the cell.”
Shin and postdoctoral fellows Fuguo Jiang and Jun-Jie Liu are the three first authors of the paper, which was also co-authored by Benjamin Rauch of UCSF and postdoc Nicolas Bray, researcher Seung Hyun Baik and professor Eva Nogales, in addition to Corn and Doudna, of IGI and UC Berkeley’s Department of Molecular and Cell Biology. Doudna and Nogales are Howard Hughes Medical Institute investigators.
The work was supported in part by HHMI, the Li Ka Shing Foundation, the Heritage Medical Research Institute and the National Institute on Aging.
By Robert Sanders, Media relations


